PoSyMed

Population Systems Medicine for De Novo Mechanotyping of Complex Diseases - Subtypes of Brain ageing as an Example
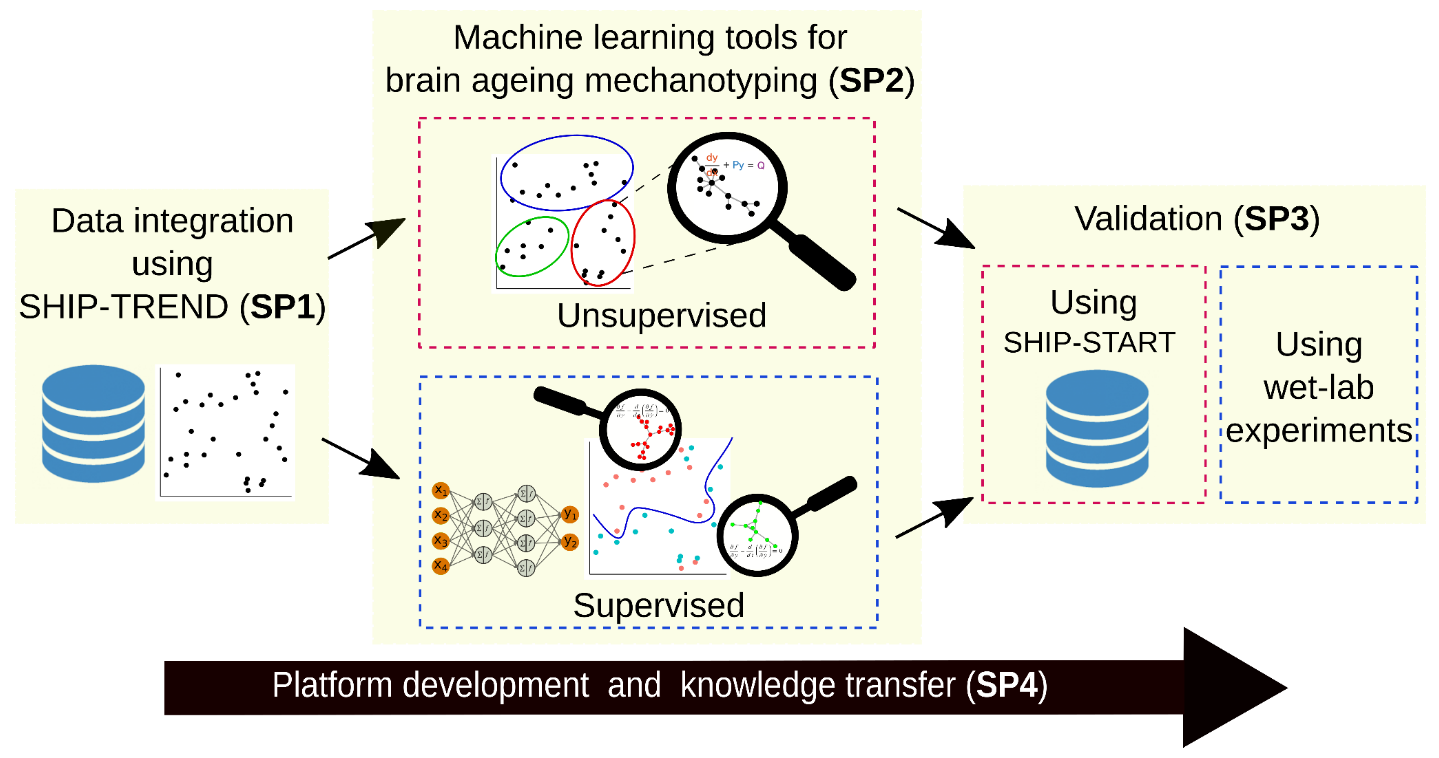
PoSyMed will analize population level data of the Study of Health in Pomerania (SHIP) in Greifswald, which has been set up as a prospective population-based study comprising two cohorts (SHIP-START and SHIP-TREND). SHIP was designed to assess the prevalence of common risk factors and diseases, and to investigate their associations.
We will develop novel unsupervised and supervised machine learning approaches for integrative disease mechanotyping using cognitive decline on a population scale as an example, where we will integrate multi-omics data from the SHIP-TREND cohort to train our machine learning models and to predict mechanistic biomarkers that stratify the underlying subtypes (de novo or by using additional clinical parameters). Subsequently, the predicted biomarker candidates will be validated, first internally using existing data and subsequently on external, new data from the independent SHIP-START cohort.
Useful links
- DESMOND
Zolotareva, Olga et al. “Identification of differentially expressed gene modules in heterogeneous diseases.” Bioinformatics (Oxford, England) vol. 37,12 (2021): 1691-1698. doi:10.1093/bioinformatics/btaa1038 - BiCoN
Lazareva, Olga et al. “BiCoN: network-constrained biclustering of patients and omics data.” Bioinformatics (Oxford, England) vol. 37,16 (2021): 2398-2404. doi:10.1093/bioinformatics/btaa1076 - MosBi
Rose, Tim Daniel et al. “MoSBi: Automated signature mining for molecular stratification and subtyping.” Proceedings of the National Academy of Sciences of the United States of America vol. 119,16 (2022): e2118210119. doi:10.1073/pnas.2118210119 - UnPaSt
Contacts
Prof. Dr. Jan Baumbach
University of Hamburg
Centre for Bioinformatics
Institute for Computational Systems Biology
jan.baumbach@uni-hamburg.de
Prof. Dr. Uwe Völker
University Medicine Greifswald
Interfaculty Institute for Genetics and Functional Genomics
voelker@uni-greifswald.de
Prof. Dr. Hans Grabe
University Medicine Greifswald
Department of Psychiatry and Psychotherapy
Hans.Grabe@med.uni-greifswald.de
Dr. Olga Tsoy
University of Hamburg
Centre for Bioinformatics
Institute for Computational Systems Biology
olga.tsoy@uni-hamburg.de
Prof. Dr. Peter R. Mertens
Otto-von-Guericke University Magdeburg
Clinic for Nephrology, Hypertension, Diabetes and Endocrinology
peter.mertens@med.ovgu.de